islr notes and exercises from An Introduction to Statistical Learning
10. Unsupervised Learning
Exercise 1: Part of the argument that the -means clustering algorithm 10.1 reaches a local optimum
- Exercise 1: Part of the argument that the -means clustering algorithm 10.1 reaches a local optimum
- a. Proving identity (10.12)
- b. Arguing the algorithm decreases the objective
- Exercise 2: Sketching a dendrogram
- a. Complete linkage dendrogram
- b. Single linkage dendrogram
- c. Clusters from the complete linkage dendrogram
- d.
- e.
- Exercise 3: Manual example of -means clustering for , , .
- Exercise 4: Comparing single and complete linkage
- b. For clusters ,
- Exercise 5: -means clustering in fig 10.14
- Exercise 6: A case of PCA
a. Proving identity (10.12)
We’ll prove this for the case , from which the result follows easily by induction.
Starting with the left-hand side of (10.12), we have
Now, we work with the right-hand side.
Where we have let for readability.
Furthermore
Thus (4) becomes
And (12) is the same as (2)
b. Arguing the algorithm decreases the objective
The identity (10.12) shows that the objective (10.11) is equal to
where is the -th cluster centroid.
At each iteration of the algorithm, observations are reassigned to the cluster whose centroid is closest, i.e. such that is minimal over . That is, if denotes the cluster assigned to observation on iteration , and the centroid of cluster then
for all . Thus (1) decreases at each iteration.
Exercise 2: Sketching a dendrogram
a. Complete linkage dendrogram
%matplotlib inline
from matplotlib import pyplot as plt
from scipy.cluster.hierarchy import dendrogram, linkage
from scipy.spatial.distance import squareform
import numpy as np
dist_sq = np.array([[0, 0.3, 0.4, 0.7], [0.3, 0, 0.5, 0.8],
[0.4, 0.5, 0, 0.45], [0.7, 0.8, 0.45, 0]])
y = squareform(dist_sq)
Z = linkage(y, method='complete', metric='Euclidean')
plt.title('Complete Linkage Dendrogram')
plt.xlabel('sample')
plt.ylabel('Euclidean Distance')
dendrogram(Z, labels=['x1', 'x2', 'x3', 'x4'], leaf_rotation=45)
{'icoord': [[5.0, 5.0, 15.0, 15.0],
[25.0, 25.0, 35.0, 35.0],
[10.0, 10.0, 30.0, 30.0]],
'dcoord': [[0.0, 0.3, 0.3, 0.0],
[0.0, 0.45, 0.45, 0.0],
[0.3, 0.8, 0.8, 0.45]],
'ivl': ['x1', 'x2', 'x3', 'x4'],
'leaves': [0, 1, 2, 3],
'color_list': ['g', 'r', 'b']}

b. Single linkage dendrogram
Z = linkage(y, method='single', metric='Euclidean')
plt.title('Single Linkage Dendrogram')
plt.xlabel('sample')
plt.ylabel('Euclidean Distance')
dendrogram(Z, labels=['x1', 'x2', 'x3', 'x4'], leaf_rotation=90)
{'icoord': [[25.0, 25.0, 35.0, 35.0],
[15.0, 15.0, 30.0, 30.0],
[5.0, 5.0, 22.5, 22.5]],
'dcoord': [[0.0, 0.3, 0.3, 0.0],
[0.0, 0.4, 0.4, 0.3],
[0.0, 0.45, 0.45, 0.4]],
'ivl': ['x4', 'x3', 'x1', 'x2'],
'leaves': [3, 2, 0, 1],
'color_list': ['g', 'b', 'b']}
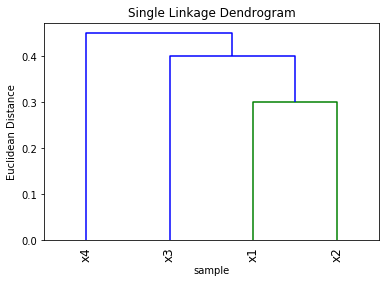
c. Clusters from the complete linkage dendrogram
The clusters are
d.
The clusters are
e.
Just exchange in the diagram in part a.
Exercise 3: Manual example of -means clustering for , , .
import pandas as pd
import seaborn as sns; sns.set_style('whitegrid')
df = pd.DataFrame({'X1': [1, 1, 0, 5, 6, 4], 'X2': [4, 3, 4, 1, 2, 0]},
index=range(1, 7))
df
| X1 | X2 | |
|---|---|---|
| 1 | 1 | 4 |
| 2 | 1 | 3 |
| 3 | 0 | 4 |
| 4 | 5 | 1 |
| 5 | 6 | 2 |
| 6 | 4 | 0 |
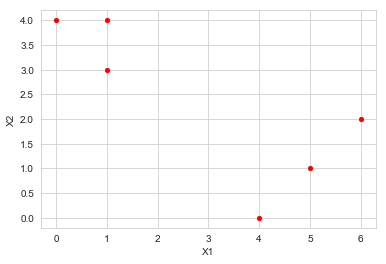
a. Plot the observations
sns.scatterplot(x='X1', y='X2', data=df, color='r')
<matplotlib.axes._subplots.AxesSubplot at 0x8168b3d30>
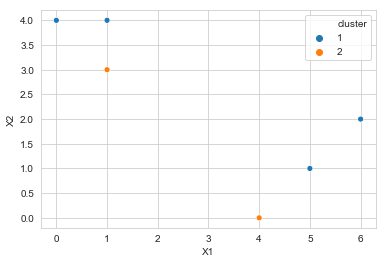
b. Initialize with random cluster assignment
np.random.seed(33)
df['cluster'] = np.random.choice([1, 2], replace=True, size=6)
df['cluster']
1 1
2 2
3 1
4 1
5 1
6 2
Name: cluster, dtype: int64
sns.scatterplot(x='X1', y='X2', data=df, hue='cluster', legend='full',
palette=sns.color_palette(n_colors=2))
<matplotlib.axes._subplots.AxesSubplot at 0x8168bad68>
c. Compute the centroid for each cluster
def get_centroids(df):
# compute centroids
c1, c2 = df[df['cluster'] == 1], df[df['cluster'] == 2]
cent_1 = [c1['X1'].mean(), c1['X2'].mean()]
cent_2 = [c2['X1'].mean(), c2['X2'].mean()]
return (cent_1, cent_2)
cent_1, cent_2 = get_centroids(df)
d. Assign observations to clusters by centroid
def d_cent(cent):
def f(x):
return np.linalg.norm(x - cent)
return f
def assign_to_centroid(cent_1, cent_2):
def f(x):
d_1, d_2 = d_cent(cent_1)(x), d_cent(cent_2)(x)
return 1 if d_1 < d_2 else 2
return f
def assign_to_clusters(df):
cent_1, cent_2 = get_centroids(df)
df = df.drop(columns=['cluster'])
return df.apply(assign_to_centroid(cent_1, cent_2), axis=1)
assign_to_clusters(df)
1 1
2 1
3 1
4 2
5 1
6 2
dtype: int64
e. Iterate c., d. until cluster assignments are stable
def get_final_clusters(df):
cl_before, cl_after = df['cluster'], assign_to_clusters(df)
while not (cl_before == cl_after).any():
df.loc[:, 'cluster'] = cl_after
get_final_clusters(df)
return cl_after
get_final_clusters(df)
1 1
2 1
3 1
4 2
5 1
6 2
dtype: int64
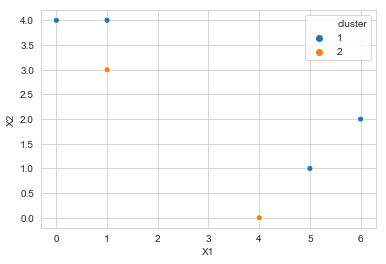
f. Plot clusters
sns.scatterplot(x='X1', y='X2', data=df, hue='cluster', legend='full',
palette=sns.color_palette(n_colors=2))
<matplotlib.axes._subplots.AxesSubplot at 0x1a191f0198>
Exercise 4: Comparing single and complete linkage
a. For clusters and .
In both linkage diagrams, the clusters and fuse when they are the most similar pair of clusters among all pairs at that height.
In the simple linkage diagram, this occurs at the height when the minimum dissimilarity over pairs is less than for any other clusters . In the complete linkage diagram, this occurs at the height when the maximum dissimilarity over pairs is less than for any other clusters .
It is possible to have the maximum over other clusters less than or equal to the minimum, and vice versa. In the former case, and in the latter, . So there is not enough information to tell.
To make this argument a bit more concrete here are some examples:
An example of
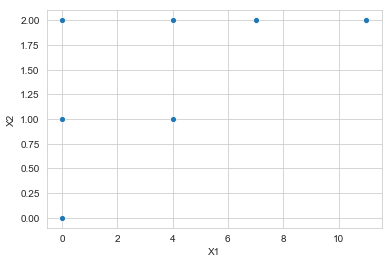
Consider the following possible subset of observations. Suppose all other observations are far away.
array = np.array([[0, 0], [0, 1], [0, 2],
[4, 1], [4, 2],
[7, 2], [11, 2]])
df1 = pd.DataFrame(array, columns=['X1', 'X2'], index=range(1, 8))
df1
| X1 | X2 | |
|---|---|---|
| 1 | 0 | 0 |
| 2 | 0 | 1 |
| 3 | 0 | 2 |
| 4 | 4 | 1 |
| 5 | 4 | 2 |
| 6 | 7 | 2 |
| 7 | 11 | 2 |
sns.scatterplot(x='X1', y='X2', data=df1)
<matplotlib.axes._subplots.AxesSubplot at 0x1a1b1abc88>
fig, ax = plt.subplots(1, 2, figsize=(10, 6))
# plot single linkage dendrogram
plt.subplot(1, 2, 1)
Z = linkage(df1[['X1', 'X2']], method='single', metric='Euclidean')
plt.title('Single Linkage Dendrogram')
plt.xlabel('sample')
plt.ylabel('Euclidean Distance')
dendrogram(Z, labels=['x'+stri. for i in range(1, 8)], leaf_rotation=45)
# plot complete linkage dendrogram
plt.subplot(1, 2, 2)
Z = linkage(df1[['X1', 'X2']], method='complete', metric='Euclidean')
plt.title('Complete Linkage Dendrogram')
plt.xlabel('sample')
plt.ylabel('Euclidean Distance')
dendrogram(Z, labels=['x'+stri. for i in range(1, 8)], leaf_rotation=45)
fig.tight_layout()
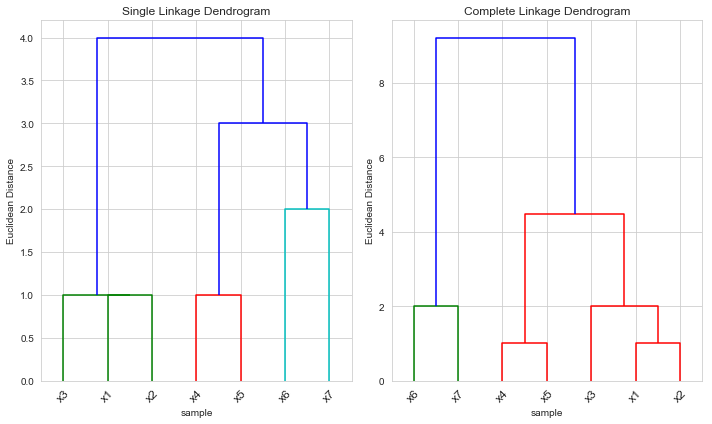
In this example, and are indeed both clusters – they are formed at height 1 in the simple linkage dendrogram and at height 2 in the complete linkage dendrogram.
But in this case the height that they are fused is greater for the complete linkage than for the single linkage
An example of
We use the same observations from the last example, except the point changes
df1.loc[7, 'X1'] = 9
df1
| X1 | X2 | |
|---|---|---|
| 1 | 0 | 0 |
| 2 | 0 | 1 |
| 3 | 0 | 2 |
| 4 | 4 | 1 |
| 5 | 4 | 2 |
| 6 | 7 | 2 |
| 7 | 9 | 2 |
fig, ax = plt.subplots(1, 2, figsize=(10, 6))
# plot single linkage dendrogram
plt.subplot(1, 2, 1)
Z = linkage(df1[['X1', 'X2']], method='single', metric='Euclidean')
plt.title('Single Linkage Dendrogram')
plt.xlabel('sample')
plt.ylabel('Euclidean Distance')
dendrogram(Z, labels=['x'+stri. for i in range(1, 8)], leaf_rotation=45)
# plot complete linkage dendrogram
plt.subplot(1, 2, 2)
Z = linkage(df1[['X1', 'X2']], method='complete', metric='Euclidean')
plt.title('Complete Linkage Dendrogram')
plt.xlabel('sample')
plt.ylabel('Euclidean Distance')
dendrogram(Z, labels=['x'+stri. for i in range(1, 8)], leaf_rotation=45)
fig.tight_layout()
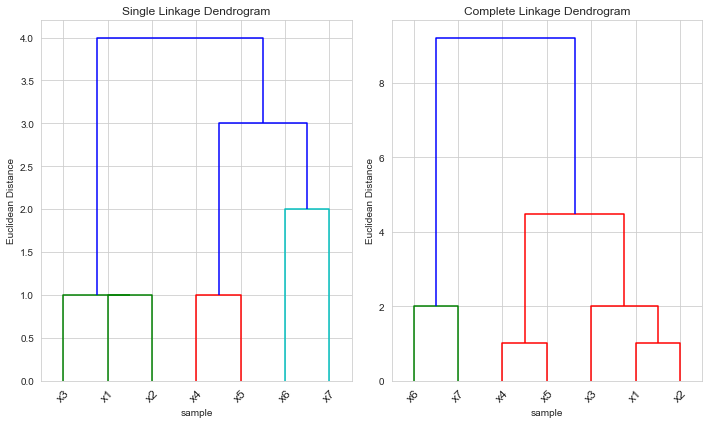
Again in this example, and are indeed both clusters – again they are formed at height 1 in the simple linkage dendrogram and at height 2 in the complete linkage dendrogram.
But in this case the height that they are fused is greater for the single linkage than for the complete linkage
b. For clusters ,
In this case, with singleton clusters ,
so are fused when in the single linkage diagram when
and in the complete linkage diagram when
so necessarily
Exercise 5: -means clustering in fig 10.14
For the left-hand scaling, we would expect the orange customer in a cluster by themselves, and the other 7 customers in the other. The orange customer has a minimum distance of 3 from any other customer, and all other 7 customers are a distance one away from some other customer in that group.
For the middle scaling, we would expect to see the customers that bought computers (yellow, blue, red, magenta) in one cluster and the others in the other.
For the right-hand scaling, we would expect to see the same as for the middle scaling.
We can verify these expectations with a computation
Left-hand scaling
customers = ['black', 'orange', 'lt_blue', 'green', 'yellow', 'dk_blue',
'red', 'magenta']
purchases = [[8, 0], [11, 0], [7, 0], [6, 0], [5, 1], [6, 1], [7, 1],
[8, 1]]
df_left = pd.DataFrame(purchases, columns=['socks', 'computers'], index=customers)
df_left
| socks | computers | |
|---|---|---|
| black | 8 | 0 |
| orange | 11 | 0 |
| lt_blue | 7 | 0 |
| green | 6 | 0 |
| yellow | 5 | 1 |
| dk_blue | 6 | 1 |
| red | 7 | 1 |
| magenta | 8 | 1 |
fig, ax = plt.subplots(1, 2, figsize=(10, 6))
# plot single linkage dendrogram
plt.subplot(1, 2, 1)
Z = linkage(df_left, method='single', metric='Euclidean')
plt.title('Single Linkage Dendrogram')
plt.xlabel('sample')
plt.ylabel('Euclidean Distance')
dendrogram(Z, labels=customers, leaf_rotation=45)
# plot complete linkage dendrogram
plt.subplot(1, 2, 2)
Z = linkage(df_left, method='complete', metric='Euclidean')
plt.title('Complete Linkage Dendrogram')
plt.xlabel('sample')
plt.ylabel('Euclidean Distance')
dendrogram(Z, labels=customers, leaf_rotation=45)
fig.tight_layout()
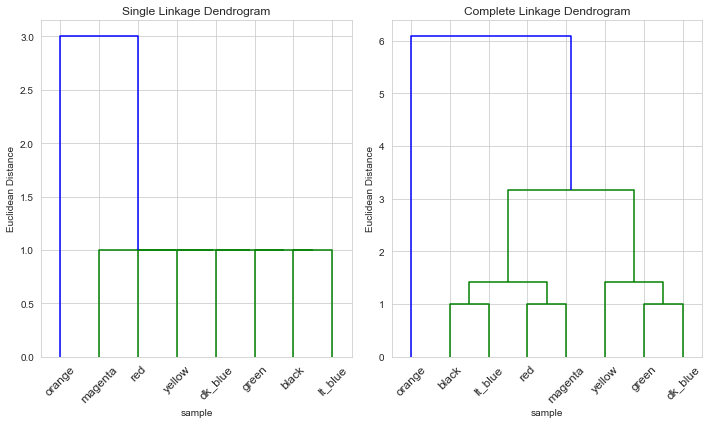
For both single and complete linkages, the clusters were as expected
Middle scaling
We don’t get the same plot as the book whether we scale by the standard deviation (or the variance) so we’ll code this by hand again (with a bit of guesswork)
df_mid = df/df.std()
df_mid.loc[:, 'socks'] = [1.0, 1.4, 0.9, 0.78, 0.62, 0.78, 0.9, 1.0]
df_mid.loc[:, 'computers'] = [0, 0, 0, 0, 1.4, 1.4, 1.4, 1.4]
df_mid
| socks | computers | |
|---|---|---|
| black | 1.00 | 0.0 |
| orange | 1.40 | 0.0 |
| lt_blue | 0.90 | 0.0 |
| green | 0.78 | 0.0 |
| yellow | 0.62 | 1.4 |
| dk_blue | 0.78 | 1.4 |
| red | 0.90 | 1.4 |
| magenta | 1.00 | 1.4 |
fig, ax = plt.subplots(1, 2, figsize=(10, 6))
# plot single linkage dendrogram
plt.subplot(1, 2, 1)
Z = linkage(df_mid, method='single', metric='Euclidean')
plt.title('Single Linkage Dendrogram')
plt.xlabel('sample')
plt.ylabel('Euclidean Distance')
dendrogram(Z, labels=customers, leaf_rotation=45)
# plot complete linkage dendrogram
plt.subplot(1, 2, 2)
Z = linkage(df_mid, method='complete', metric='Euclidean')
plt.title('Complete Linkage Dendrogram')
plt.xlabel('sample')
plt.ylabel('Euclidean Distance')
dendrogram(Z, labels=customers, leaf_rotation=45)
fig.tight_layout()
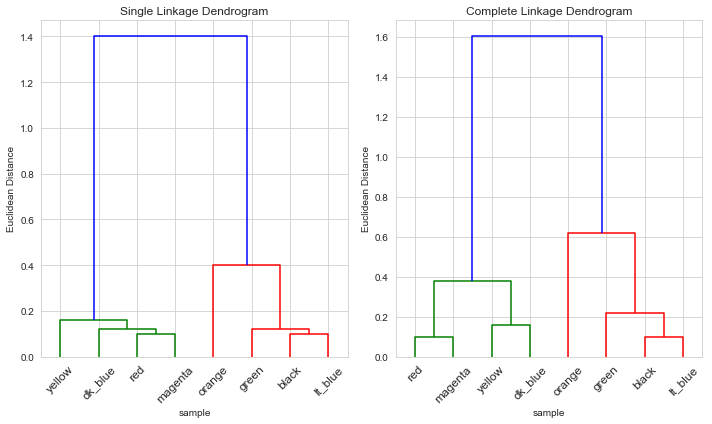
For both single and complete linkages, the clusters were again as expected
Right-hand scaling
We’ll assume per pair of socks, and per computer
df_right = df_left.copy()
df_right.loc[:, 'socks'] = 2 * df_right['socks']
df_right.loc[:, 'computers'] = 2000 * df_right['computers']
df_right
| socks | computers | |
|---|---|---|
| black | 16 | 0 |
| orange | 22 | 0 |
| lt_blue | 14 | 0 |
| green | 12 | 0 |
| yellow | 10 | 2000 |
| dk_blue | 12 | 2000 |
| red | 14 | 2000 |
| magenta | 16 | 2000 |
fig, ax = plt.subplots(1, 2, figsize=(10, 6))
# plot single linkage dendrogram
plt.subplot(1, 2, 1)
Z = linkage(df_right, method='single', metric='Euclidean')
plt.title('Single Linkage Dendrogram')
plt.xlabel('sample')
plt.ylabel('Euclidean Distance')
dendrogram(Z, labels=customers, leaf_rotation=45)
# plot complete linkage dendrogram
plt.subplot(1, 2, 2)
Z = linkage(df_right, method='complete', metric='Euclidean')
plt.title('Complete Linkage Dendrogram')
plt.xlabel('sample')
plt.ylabel('Euclidean Distance')
dendrogram(Z, labels=customers, leaf_rotation=45)
fig.tight_layout()
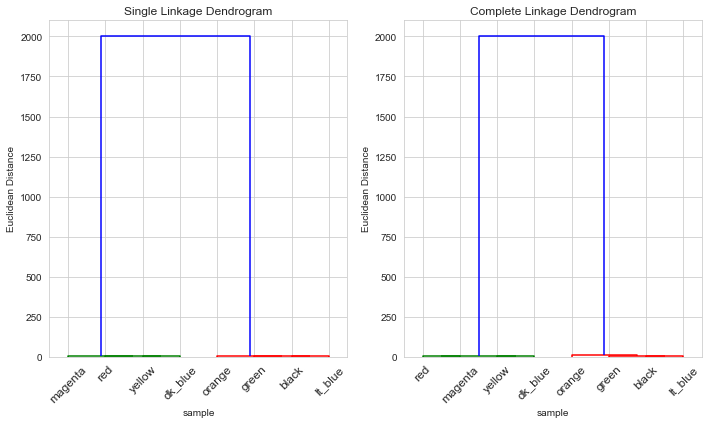
Again for both single and complete linkages, the clusters were as expected
Exercise 6: A case of PCA
a.
This means that the proportion of variance explained by the first principal component is 0.10. That is, the ratio of the (sample) variance of the first component to the total (sample) variance is 0.10