islr notes and exercises from An Introduction to Statistical Learning
7. Moving Beyond Linearity
Exercise 7: Using non-linear multiple regression to predict wage in Wage dataset
We’re modifying the exercise a bit to consider multiple regression (as opposed to considering different predictors individually). It’s not hard to see how the techniques of this chapter generalize to the multiple regression setting.
Preparing the data
Loading
%matplotlib inline
import numpy as np
import pandas as pd
import seaborn as sns; sns.set()
wage = pd.read_csv("../../datasets/Wage.csv")
wage.head()
| Unnamed: 0 | year | age | maritl | race | education | region | jobclass | health | health_ins | logwage | wage | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 231655 | 2006 | 18 | 1. Never Married | 1. White | 1. < HS Grad | 2. Middle Atlantic | 1. Industrial | 1. <=Good | 2. No | 4.318063 | 75.043154 |
| 1 | 86582 | 2004 | 24 | 1. Never Married | 1. White | 4. College Grad | 2. Middle Atlantic | 2. Information | 2. >=Very Good | 2. No | 4.255273 | 70.476020 |
| 2 | 161300 | 2003 | 45 | 2. Married | 1. White | 3. Some College | 2. Middle Atlantic | 1. Industrial | 1. <=Good | 1. Yes | 4.875061 | 130.982177 |
| 3 | 155159 | 2003 | 43 | 2. Married | 3. Asian | 4. College Grad | 2. Middle Atlantic | 2. Information | 2. >=Very Good | 1. Yes | 5.041393 | 154.685293 |
| 4 | 11443 | 2005 | 50 | 4. Divorced | 1. White | 2. HS Grad | 2. Middle Atlantic | 2. Information | 1. <=Good | 1. Yes | 4.318063 | 75.043154 |
wage.info()
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 3000 entries, 0 to 2999
Data columns (total 12 columns):
Unnamed: 0 3000 non-null int64
year 3000 non-null int64
age 3000 non-null int64
maritl 3000 non-null object
race 3000 non-null object
education 3000 non-null object
region 3000 non-null object
jobclass 3000 non-null object
health 3000 non-null object
health_ins 3000 non-null object
logwage 3000 non-null float64
wage 3000 non-null float64
dtypes: float64(2), int64(3), object(7)
memory usage: 281.3+ KB
Cleaning
Drop columns
The unnamed column appears to be some sort of id number, which is useless for our purposes. We can also drop logwage since it’s redundant
wage = wage.drop(columns=['Unnamed: 0', 'logwage'])
Convert to numerical dtypes
wage_num = pd.get_dummies(wage)
wage_num.head()
| year | age | wage | maritl_1. Never Married | maritl_2. Married | maritl_3. Widowed | maritl_4. Divorced | maritl_5. Separated | race_1. White | race_2. Black | ... | education_3. Some College | education_4. College Grad | education_5. Advanced Degree | region_2. Middle Atlantic | jobclass_1. Industrial | jobclass_2. Information | health_1. <=Good | health_2. >=Very Good | health_ins_1. Yes | health_ins_2. No | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 2006 | 18 | 75.043154 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | ... | 0 | 0 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 1 |
| 1 | 2004 | 24 | 70.476020 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | ... | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 1 |
| 2 | 2003 | 45 | 130.982177 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | ... | 1 | 0 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 0 |
| 3 | 2003 | 43 | 154.685293 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 1 | 0 |
| 4 | 2005 | 50 | 75.043154 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | ... | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 0 |
5 rows × 24 columns
Preprocessing
Scaling the numerical variables
df = wage_num[['year', 'age', 'wage']]
wage_num_std = wage_num.copy()
wage_num_std.loc[:, ['year', 'age', 'wage']] = (df - df.mean())/df.std()
wage_num_std.head()
| year | age | wage | maritl_1. Never Married | maritl_2. Married | maritl_3. Widowed | maritl_4. Divorced | maritl_5. Separated | race_1. White | race_2. Black | ... | education_3. Some College | education_4. College Grad | education_5. Advanced Degree | region_2. Middle Atlantic | jobclass_1. Industrial | jobclass_2. Information | health_1. <=Good | health_2. >=Very Good | health_ins_1. Yes | health_ins_2. No | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0.103150 | -2.115215 | -0.878545 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | ... | 0 | 0 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 1 |
| 1 | -0.883935 | -1.595392 | -0.987994 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | ... | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 1 |
| 2 | -1.377478 | 0.223986 | 0.461999 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | ... | 1 | 0 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 0 |
| 3 | -1.377478 | 0.050712 | 1.030030 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 1 | 0 |
| 4 | -0.390392 | 0.657171 | -0.878545 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | ... | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 0 |
5 rows × 24 columns
Fitting some nonlinear models
X_sc, y_sc = wage_num_std.drop(columns=['wage']).values, wage_num_std['wage'].values
X_sc.shape, y_sc.shape
((3000, 23), (3000,))
Polynomial Ridge Regression
We don’t need a special module for this model - we can use a scikit-learn pipeline.
We’ll use 10-fold cross validation to pick the polynomial degree and L2 penalty.
from sklearn.preprocessing import PolynomialFeatures
from sklearn.pipeline import Pipeline
from sklearn.linear_model import Ridge
pr_pipe = Pipeline([('poly', PolynomialFeatures()), ('ridge', Ridge())])
pr_param_grid = dict(poly__degree=np.arange(1, 5), ridge__alpha=np.logspace(-4, 4, 5))
pr_search = GridSearchCV(pr_pipe, pr_param_grid, cv=5, scoring='neg_mean_squared_error')
pr_search.fit(X_sc, y_sc)
GridSearchCV(cv=5, error_score='raise-deprecating',
estimator=Pipeline(memory=None,
steps=[('poly', PolynomialFeatures(degree=2, include_bias=True, interaction_only=False)), ('ridge', Ridge(alpha=1.0, copy_X=True, fit_intercept=True, max_iter=None,
normalize=False, random_state=None, solver='auto', tol=0.001))]),
fit_params=None, iid='warn', n_jobs=None,
param_grid={'poly__degree': array([1, 2, 3, 4]), 'ridge__alpha': array([1.e-04, 1.e-02, 1.e+00, 1.e+02, 1.e+04])},
pre_dispatch='2*n_jobs', refit=True, return_train_score='warn',
scoring='neg_mean_squared_error', verbose=0)
pr_search.best_params_
{'poly__degree': 2, 'ridge__alpha': 100.0}
Local Regression
scikit-learn has support for local regression
from sklearn.neighbors import KNeighborsRegressor
from sklearn.model_selection import GridSearchCV
lr_param_grid = dict(n_neighbors=np.arange(1,7), weights=['uniform', 'distance'],
p=np.arange(1, 7))
lr_search = GridSearchCV(KNeighborsRegressor(), lr_param_grid, cv=10,
scoring='neg_mean_squared_error')
lr_search.fit(X_sc, y_sc)
GridSearchCV(cv=10, error_score='raise-deprecating',
estimator=KNeighborsRegressor(algorithm='auto', leaf_size=30, metric='minkowski',
metric_params=None, n_jobs=None, n_neighbors=5, p=2,
weights='uniform'),
fit_params=None, iid='warn', n_jobs=None,
param_grid={'n_neighbors': array([1, 2, 3, 4, 5, 6]), 'weights': ['uniform', 'distance'], 'p': array([1, 2, 3, 4, 5, 6])},
pre_dispatch='2*n_jobs', refit=True, return_train_score='warn',
scoring='neg_mean_squared_error', verbose=0)
lr_search.best_params_
{'n_neighbors': 6, 'p': 1, 'weights': 'uniform'}
GAMs
GAMs are quite general. There exists python modules that implement specific choices for the nonlinear component functions . Here we’ll explore two modules that seem relatively mature/well-maintained.
GAMs with pyGAM
The module pyGAM implements P-splines.
from pygam import GAM, s, f
# generate string for terms
spline_terms = ' + '.join(['s(' + stri. + ')' for i in range(0,3)])
factor_terms = ' + '.join(['f(' + stri. + ')'
for i in range(3,X_sc.shape[1])])
terms = spline_terms + ' + ' + factor_terms
terms
's(0) + s(1) + s(2) + f(3) + f(4) + f(5) + f(6) + f(7) + f(8) + f(9) + f(10) + f(11) + f(12) + f(13) + f(14) + f(15) + f(16) + f(17) + f(18) + f(19) + f(20) + f(21) + f(22)'
pygam_gam = GAM(s(0) + s(1) + s(2) + f(3) + f(4) + f(5) + f(6) + f(7)
+ f(8) + f(9) + f(10) + f(11) + f(12) + f(13) + f(14)
+ f(15) + f(16) + f(17) + f(18) + f(19) + f(20) + f(21)
+ f(22))
ps_search = pygam_gam.gridsearch(X_sc, y_sc, progress=True,
lam=np.exp(np.random.rand(100, 23) * 6 - 3))
100% (100 of 100) |######################| Elapsed Time: 0:00:13 Time: 0:00:13
Model Selection
As in exercise 6, we’ll select a model on the basis of mean squared test error.
mse_test_df = pd.DataFrame({'mse_test':np.zeros(3)}, index=['poly_ridge', 'local_reg', 'p_spline'])
# polynomial ridge and local regression models already have CV estimates of test mse
mse_test_df.at['poly_ridge', 'mse_test'] = -pr_search.best_score_
mse_test_df.at['local_reg', 'mse_test'] = -lr_search.best_score_
from sklearn.model_selection import cross_val_score
# get p-spline CV estimate of test mse
mse_test_df.at['p_spline', 'mse_test'] = -np.mean(cross_val_score(ps_search,
X_sc, y_sc, scoring='neg_mean_squared_error',
cv=10))
mse_test_df
| mse_test | |
|---|---|
| poly_ridge | 0.653614 |
| local_reg | 0.741645 |
| p_spline | 1.000513 |
Polynomial ridge regression has won out. Since this CV mse estimate was calculated on scaled data, let’s get the estimate for the original data
%%capture
X, y = wage_num.drop(columns=['wage']).values, wage_num['wage'].values
cv_score = cross_val_score(pr_search.best_estimator_, X, y, scoring='neg_mean_squared_error', cv=10)
mse = -np.mean(cv_score)
mse
1137.2123862552992
me = np.sqrt(mse)
me
33.722579768684646
sns.distplot(wage['wage'])
<matplotlib.axes._subplots.AxesSubplot at 0x1a20d1e668>
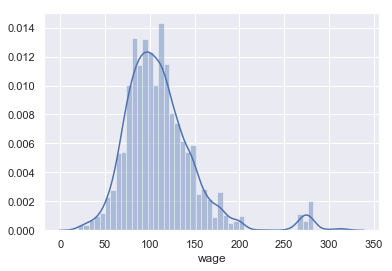
wage['wage'].describe()
count 3000.000000
mean 111.703608
std 41.728595
min 20.085537
25% 85.383940
50% 104.921507
75% 128.680488
max 318.342430
Name: wage, dtype: float64
This model predicts a mean (absolute) error of
print('{}'.format(round(me/wage['wage'].std(), 2)))
0.81
which is 0.81 standard deviations.
Improvements
After inspecting the distribution of wage, it’s fairly clear there is a group of outliers that are no doubt affecting the prediction accuracy of the model. Let’s try to separate that group.
sns.scatterplot(x=wage.index, y=wage['wage'].sort_values(), alpha=0.4, color='grey')
<matplotlib.axes._subplots.AxesSubplot at 0x1a2469d8d0>
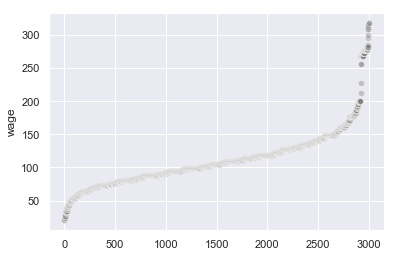
There appears to be a break point around 250. Let’s take all rows with wage less than this
wage_num_low = wage_num[wage_num['wage'] < 250]
wage_num_low_sc = wage_num_low.copy()
df = wage_num_low_sc[['year', 'age', 'wage']]
wage_num_low_sc.loc[:, ['year', 'age', 'wage']] = (df - df.mean())/df.std()
Let train the same models again
X_low_sc, y_low_sc = wage_num_low_sc.drop(columns=['wage']).values, wage_num_low_sc['wage']
# polynomial ridge model
pr_search.fit(X_low_sc, y_low_sc)
/Users/home/anaconda3/lib/python3.6/site-packages/sklearn/model_selection/_search.py:841: DeprecationWarning: The default of the `iid` parameter will change from True to False in version 0.22 and will be removed in 0.24. This will change numeric results when test-set sizes are unequal.
DeprecationWarning)
GridSearchCV(cv=5, error_score='raise-deprecating',
estimator=Pipeline(memory=None,
steps=[('poly', PolynomialFeatures(degree=2, include_bias=True, interaction_only=False)), ('ridge', Ridge(alpha=1.0, copy_X=True, fit_intercept=True, max_iter=None,
normalize=False, random_state=None, solver='auto', tol=0.001))]),
fit_params=None, iid='warn', n_jobs=None,
param_grid={'poly__degree': array([1, 2, 3, 4]), 'ridge__alpha': array([1.e-04, 1.e-02, 1.e+00, 1.e+02, 1.e+04])},
pre_dispatch='2*n_jobs', refit=True, return_train_score='warn',
scoring='neg_mean_squared_error', verbose=0)
pr_search.best_params_
{'poly__degree': 2, 'ridge__alpha': 100.0}
# local regression
lr_search.fit(X_low_sc, y_low_sc)
GridSearchCV(cv=10, error_score='raise-deprecating',
estimator=KNeighborsRegressor(algorithm='auto', leaf_size=30, metric='minkowski',
metric_params=None, n_jobs=None, n_neighbors=5, p=2,
weights='uniform'),
fit_params=None, iid='warn', n_jobs=None,
param_grid={'n_neighbors': array([1, 2, 3, 4, 5, 6]), 'weights': ['uniform', 'distance'], 'p': array([1, 2, 3, 4, 5, 6])},
pre_dispatch='2*n_jobs', refit=True, return_train_score='warn',
scoring='neg_mean_squared_error', verbose=0)
lr_search.best_params_
{'n_neighbors': 6, 'p': 5, 'weights': 'uniform'}
# p spline
ps_search = pygam_gam.gridsearch(X_low_sc, y_low_sc, progress=True,
lam=np.exp(np.random.rand(100, 23) * 6 - 3))
100% (100 of 100) |######################| Elapsed Time: 0:00:25 Time: 0:00:25
ps_search.summary()
GAM
=============================================== ==========================================================
Distribution: NormalDist Effective DoF: 28.9895
Link Function: IdentityLink Log Likelihood: -3606.1899
Number of Samples: 2921 AIC: 7272.3587
AICc: 7273.0018
GCV: 0.6157
Scale: 0.6047
Pseudo R-Squared: 0.4011
==========================================================================================================
Feature Function Lambda Rank EDoF P > x Sig. Code
================================= ==================== ============ ============ ============ ============
s(0) [13.8473] 20 7.0 1.38e-05 ***
s(1) [17.046] 20 8.3 3.52e-13 ***
s(2) [0.1439] 20 1.1 5.30e-04 ***
f(3) [11.8899] 2 1.0 3.09e-02 *
f(4) [3.5507] 2 0.9 2.96e-01
f(5) [0.1322] 2 0.9 1.79e-02 *
f(6) [14.0907] 2 0.0 3.22e-01
f(7) [8.8878] 2 1.0 2.87e-01
f(8) [0.2954] 2 1.0 3.01e-01
f(9) [2.0563] 2 0.9 6.03e-01
f(10) [8.6475] 2 0.0 3.51e-01
f(11) [8.7695] 2 1.0 1.11e-16 ***
f(12) [0.1363] 2 1.0 2.69e-02 *
f(13) [0.9965] 2 1.0 1.11e-16 ***
f(14) [0.1233] 2 1.0 1.11e-16 ***
f(15) [0.3011] 2 0.0 1.11e-16 ***
f(16) [2.094] 1 0.0 1.00e+00
f(17) [0.6317] 2 1.0 2.06e-01
f(18) [16.422] 2 0.0 2.07e-01
f(19) [0.3865] 2 1.0 1.33e-05 ***
f(20) [4.8438] 2 0.0 1.38e-05 ***
f(21) [0.1626] 2 1.0 1.11e-16 ***
f(22) [0.3707] 2 0.0 1.11e-16 ***
intercept 1 0.0 2.88e-01
==========================================================================================================
Significance codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
WARNING: Fitting splines and a linear function to a feature introduces a model identifiability problem
which can cause p-values to appear significant when they are not.
WARNING: p-values calculated in this manner behave correctly for un-penalized models or models with
known smoothing parameters, but when smoothing parameters have been estimated, the p-values
are typically lower than they should be, meaning that the tests reject the null too readily.
/Users/home/anaconda3/lib/python3.6/site-packages/ipykernel_launcher.py:1: UserWarning: KNOWN BUG: p-values computed in this summary are likely much smaller than they should be.
Please do not make inferences based on these values!
Collaborate on a solution, and stay up to date at:
github.com/dswah/pyGAM/issues/163
"""Entry point for launching an IPython kernel.
low_mse_test_df = pd.DataFrame({'low_mse_test':np.zeros(3)}, index=['poly_ridge', 'local_reg', 'p_spline'])
low_mse_test_df.at['poly_ridge', 'low_mse_test'] = -pr_search.best_score_
low_mse_test_df.at['local_reg', 'low_mse_test'] = -lr_search.best_score_
low_mse_test_df.at['p_spline', 'low_mse_test'] = -np.mean(cross_val_score(ps_search,
X_low_sc, y_low_sc, scoring='neg_mean_squared_error',
cv=10))
mse_df = pd.concat([mse_test_df, low_mse_test_df], axis=1)
mse_df
| mse_test | low_mse_test | |
|---|---|---|
| poly_ridge | 0.653614 | 0.613098 |
| local_reg | 0.741645 | 0.701615 |
| p_spline | 1.000513 | 1.000513 |
There was a considerable improvement for the polynomial ridge and local regression models.