islr notes and exercises from An Introduction to Statistical Learning
6. Linear Model Selection and Regularization
Exercise 9: Predicting Apps in College dataset
Preparing the data
import pandas as pd
college = pd.read_csv('../../datasets/College.csv')
college = college.rename({'Unnamed: 0': 'Name'}, axis='columns')
college.head()
| Name | Private | Apps | Accept | Enroll | Top10perc | Top25perc | F.Undergrad | P.Undergrad | Outstate | Room.Board | Books | Personal | PhD | Terminal | S.F.Ratio | perc.alumni | Expend | Grad.Rate | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | Abilene Christian University | Yes | 1660 | 1232 | 721 | 23 | 52 | 2885 | 537 | 7440 | 3300 | 450 | 2200 | 70 | 78 | 18.1 | 12 | 7041 | 60 |
| 1 | Adelphi University | Yes | 2186 | 1924 | 512 | 16 | 29 | 2683 | 1227 | 12280 | 6450 | 750 | 1500 | 29 | 30 | 12.2 | 16 | 10527 | 56 |
| 2 | Adrian College | Yes | 1428 | 1097 | 336 | 22 | 50 | 1036 | 99 | 11250 | 3750 | 400 | 1165 | 53 | 66 | 12.9 | 30 | 8735 | 54 |
| 3 | Agnes Scott College | Yes | 417 | 349 | 137 | 60 | 89 | 510 | 63 | 12960 | 5450 | 450 | 875 | 92 | 97 | 7.7 | 37 | 19016 | 59 |
| 4 | Alaska Pacific University | Yes | 193 | 146 | 55 | 16 | 44 | 249 | 869 | 7560 | 4120 | 800 | 1500 | 76 | 72 | 11.9 | 2 | 10922 | 15 |
college.loc[:, 'Private'] = [0 if entry == 'No' else 1 for entry in college['Private']]
college.info()
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 777 entries, 0 to 776
Data columns (total 19 columns):
Name 777 non-null object
Private 777 non-null int64
Apps 777 non-null int64
Accept 777 non-null int64
Enroll 777 non-null int64
Top10perc 777 non-null int64
Top25perc 777 non-null int64
F.Undergrad 777 non-null int64
P.Undergrad 777 non-null int64
Outstate 777 non-null int64
Room.Board 777 non-null int64
Books 777 non-null int64
Personal 777 non-null int64
PhD 777 non-null int64
Terminal 777 non-null int64
S.F.Ratio 777 non-null float64
perc.alumni 777 non-null int64
Expend 777 non-null int64
Grad.Rate 777 non-null int64
dtypes: float64(1), int64(17), object(1)
memory usage: 115.4+ KB
a. Train - test split
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(college.drop(columns=['Apps', 'Name']),
college['Apps'])
b. Linear regression model
from sklearn.linear_model import LinearRegression
from sklearn.metrics import mean_squared_error
linreg = LinearRegression().fit(X_train, y_train)
linreg_mse_test = mean_squared_error(y_test, linreg.predict(X_test))
mses_df = pd.DataFrame({'mse_test': linreg_mse_test},
index=['linreg'])
mses_df
| mse_test | |
|---|---|
| linreg | 1.869641e+06 |
c. Ridge regression model
from sklearn.linear_model import Ridge
from sklearn.model_selection import GridSearchCV
parameters = {'alpha': [10**i for i in range(-3, 4)]}
ridge = GridSearchCV(Ridge(), parameters, cv=10,
scoring='neg_mean_squared_error')
%%capture
ridge.fit(X_train, y_train)
%%capture
ridge_cv_df = pd.DataFrame(ridge.cv_results_)
ridge_cv_df
ridge_mse_test = mean_squared_error(y_test, ridge.best_estimator_.predict(X_test))
mses_df = mses_df.append(pd.DataFrame({'mse_test': ridge_mse_test}, index=['ridge']))
mses_df
| mse_test | |
|---|---|
| linreg | 1.869641e+06 |
| ridge | 1.875181e+06 |
d. Lasso regression model
from sklearn.linear_model import Lasso
from sklearn.model_selection import GridSearchCV
parameters = {'alpha': [10**i for i in range(-3, 4)]}
lasso = GridSearchCV(Lasso(), parameters, cv=10, scoring='neg_mean_squared_error')
%%capture
lasso.fit(X_train, y_train)
%%capture
lasso_cv_df = pd.DataFrame(lasso.cv_results_)
lasso_cv_df
lasso_mse_test = mean_squared_error(y_test, lasso.best_estimator_.predict(X_test))
mses_df = mses_df.append(pd.DataFrame({'mse_test': lasso_mse_test}, index=['lasso']))
mses_df
| mse_test | |
|---|---|
| linreg | 1.869641e+06 |
| ridge | 1.875181e+06 |
| lasso | 1.870846e+06 |
e. PCR model
scikit-learn doesn’t have combined PCA and regression so we’ll use the top answer to this CrossValidated question
import numpy as np
from sklearn.preprocessing import scale
from sklearn.decomposition import PCA
from sklearn.model_selection import cross_val_score
n = len(X_train_reduced)
linreg = LinearRegression()
pcr_mses = [-cross_val_score(linreg, np.ones((n,1)), y_train, cv=10,
scoring='neg_mean_squared_error').mean()]
for i in range(1, college.shape[1] - b1):
pcr_mses += [-cross_val_score(linreg, X_train_reduced[:, :i], y_train, cv=10,
scoring='neg_mean_squared_error').mean()]
np.argmin(pcr_mses)
17
10 fold Cross-validation selects (full PCR model with no intercept).
pcr = LinearRegression().fit(X_train.iloc[:, :np.argmin(pcr_mses)], y_train)
The test error of this model is
pcr_mse_test = mean_squared_error(y_test, pcr.predict(X_test))
mses_df = mses_df.append(pd.DataFrame({'mse_test': pcr_mse_test}, index=['pcr']))
mses_df
| mse_test | |
|---|---|
| linreg | 1.869641e+06 |
| ridge | 1.875181e+06 |
| lasso | 1.870846e+06 |
| pcr | 1.869641e+06 |
f. PLS model
from sklearn.cross_decomposition import PLSRegression
# mse for only constant predictor same as for pcr
pls_mses = pcr_mses[:1]
for i in range(1, college.shape[1] - 1):
pls_mses += [-cross_val_score(estimator=PLSRegression(n_components = i),
X=X_train, y=y_train, cv=10,
scoring='neg_mean_squared_error').mean()]
np.argmin(pls_mses)
13
10 fold CV selects
pls = PLSRegression(n_components=13).fit(X_train, y_train)
pls_mse_test = mean_squared_error(y_test, pls.predict(X_test))
mses_df = mses_df.append(pd.DataFrame({'mse_test': pls_mse_test}, index=['pls']))
mses_df
| mse_test | |
|---|---|
| linreg | 1.869641e+06 |
| ridge | 1.875181e+06 |
| lasso | 1.870846e+06 |
| pcr | 1.869641e+06 |
| pls | 1.862860e+06 |
g. Comments
How accurately can we predict applications?
The test mses for each model were . This corresponds to an (absolute) error of . Given the distribution of applications
college['Apps'].describe()
count 777.000000
mean 3001.638353
std 3870.201484
min 81.000000
25% 776.000000
50% 1558.000000
75% 3624.000000
max 48094.000000
Name: Apps, dtype: float64
import seaborn as sns
% matplotlib inline
sns.distplot(college['Apps'])
<matplotlib.axes._subplots.AxesSubplot at 0x1a212479b0>
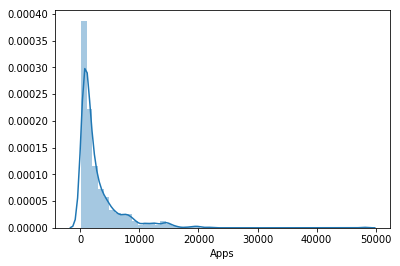
The prediction doesn’t seem that accurate. Given that distribution is highly concentrated about the mean and the upper quartile is , we can say that for most values, the prediction is off by of the true value.
Is there much difference among the test errors
No