islr notes and exercises from An Introduction to Statistical Learning
5. Resampling Methods
Exercise 8: Cross-validation on simulated data
a. Generate data
import numpy as np
np.random.seed(0)
X = np.random.normal(size=100)
Y = -2*X**2 + X + np.random.normal(size=100)
The model here is
Where .
The sample size is . Since we don’t know polynomial regression yet, we have , i.e. .
b. Scatter plot
import seaborn as sns
%matplotlib inline
sns.scatterplot(X, Y)
<matplotlib.axes._subplots.AxesSubplot at 0x1a1f70d4e0>
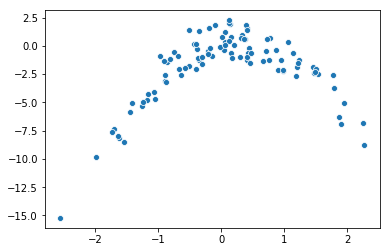
c. LOOCV errors for various models
Dataframe
import pandas as pd
data = pd.DataFrame({'const': len(X)*[1], 'X': X, 'Y': Y})
for i in range(2,5):
data['X_' + stri.] = X**i
data.head()
| const | X | Y | X_2 | X_3 | X_4 | |
|---|---|---|---|---|---|---|
| 0 | 1 | 1.764052 | -2.576558 | 3.111881 | 5.489520 | 9.683801 |
| 1 | 1 | 0.400157 | -1.267853 | 0.160126 | 0.064075 | 0.025640 |
| 2 | 1 | 0.978738 | -2.207603 | 0.957928 | 0.937561 | 0.917626 |
| 3 | 1 | 2.240893 | -6.832915 | 5.021602 | 11.252875 | 25.216490 |
| 4 | 1 | 1.867558 | -6.281111 | 3.487773 | 6.513618 | 12.164559 |
Models
from sklearn.linear_model import LinearRegression
models = {}
models['deg1'] = LinearRegression(data[['const', 'X']], data['Y'])
models['deg2'] = LinearRegression(data[['const', 'X', 'X_2']], data['Y'])
models['deg3'] = LinearRegression(data[['const', 'X', 'X_2', 'X_3']], data['Y'])
models['deg4'] = LinearRegression(data[['const', 'X', 'X_2', 'X_3', 'X_4']], data['Y'])
LOOCV errors
from sklearn.model_selection import LeaveOneOut
loocv = LeaveOneOut()
errors = []
### Degree 1 model
X = data[['const', 'X']].values
y = data['Y'].values
y_pred = np.array([])
for train_index, test_index in loocv.split(X):
X_train, X_test, y_train, y_test = X[train_index], X[test_index], y[train_index], y[test_index]
y_pred = np.append(y_pred, LinearRegression().fit(X_train, y_train).predict(X_test))
errors += [abs(y-y_pred).mean()]
### Degree 2 model
X = data[['const', 'X', 'X_2']].values
y = data['Y'].values
y_pred = np.array([])
for train_index, test_index in loocv.split(X):
X_train, X_test, y_train, y_test = X[train_index], X[test_index], y[train_index], y[test_index]
y_pred = np.append(y_pred, LinearRegression().fit(X_train, y_train).predict(X_test))
errors += [abs(y-y_pred).mean()]
### Degree 3 model
X = data[['const', 'X', 'X_2', 'X_3']].values
y = data['Y'].values
y_pred = np.array([])
for train_index, test_index in loocv.split(X):
X_train, X_test, y_train, y_test = X[train_index], X[test_index], y[train_index], y[test_index]
y_pred = np.append(y_pred, LinearRegression().fit(X_train, y_train).predict(X_test))
errors += [abs(y-y_pred).mean()]
### Degree 4 model
X = data[['const', 'X', 'X_2', 'X_3', 'X_4']].values
y = data['Y'].values
y_pred = np.array([])
for train_index, test_index in loocv.split(X):
X_train, X_test, y_train, y_test = X[train_index], X[test_index], y[train_index], y[test_index]
y_pred = np.append(y_pred, LinearRegression().fit(X_train, y_train).predict(X_test))
errors += [abs(y-y_pred).mean()]
model_names = ['deg' + stri. for i in range(1,5)]
errors_df = pd.DataFrame({'model': model_names, 'est_LOOCV_err': errors})
errors_df
| model | est_LOOCV_err | |
|---|---|---|
| 0 | deg1 | 2.239087 |
| 1 | deg2 | 0.904460 |
| 2 | deg3 | 0.917489 |
| 3 | deg4 | 0.925485 |
d. Repeat c.
If we repeat c. we don’t get any difference, since LOOCV is deterministic.
e. Which model had the smallest error?
The degree 2 model had the smallest error. This is to be expected. Since the original data was generated by a degree 2, we expect a degree 2 model to have lower test error, and the LOOCV is an estimate of the test error
f. Hypothesis testing the coefficients
import statsmodels.formula.api as smf
smf.ols('Y ~ X', data=data).fit().summary().tables[1]
| coef | std err | t | P>|t| | [0.025 | 0.975] | |
|---|---|---|---|---|---|---|
| Intercept | -1.9487 | 0.290 | -6.726 | 0.000 | -2.524 | -1.374 |
| X | 0.8650 | 0.287 | 3.015 | 0.003 | 0.296 | 1.434 |
smf.ols('Y ~ X + X_2', data=data).fit().summary().tables[1]
| coef | std err | t | P>|t| | [0.025 | 0.975] | |
|---|---|---|---|---|---|---|
| Intercept | 0.1427 | 0.132 | 1.079 | 0.283 | -0.120 | 0.405 |
| X | 1.1230 | 0.104 | 10.829 | 0.000 | 0.917 | 1.329 |
| X_2 | -2.0668 | 0.080 | -25.700 | 0.000 | -2.226 | -1.907 |
smf.ols('Y ~ X + X_2 + X_3', data=data).fit().summary().tables[1]
| coef | std err | t | P>|t| | [0.025 | 0.975] | |
|---|---|---|---|---|---|---|
| Intercept | 0.1432 | 0.133 | 1.077 | 0.284 | -0.121 | 0.407 |
| X | 1.1626 | 0.195 | 5.975 | 0.000 | 0.776 | 1.549 |
| X_2 | -2.0668 | 0.081 | -25.575 | 0.000 | -2.227 | -1.906 |
| X_3 | -0.0148 | 0.061 | -0.240 | 0.810 | -0.137 | 0.107 |
smf.ols('Y ~ X + X_2 + X_3 + X_4', data=data).fit().summary().tables[1]
| coef | std err | t | P>|t| | [0.025 | 0.975] | |
|---|---|---|---|---|---|---|
| Intercept | 0.2399 | 0.153 | 1.563 | 0.121 | -0.065 | 0.545 |
| X | 1.1207 | 0.197 | 5.691 | 0.000 | 0.730 | 1.512 |
| X_2 | -2.3116 | 0.212 | -10.903 | 0.000 | -2.732 | -1.891 |
| X_3 | 0.0049 | 0.063 | 0.078 | 0.938 | -0.121 | 0.130 |
| X_4 | 0.0556 | 0.045 | 1.248 | 0.215 | -0.033 | 0.144 |
Observations:
- The degree 1 model fit a constant coeffient with high significance while the higher degree models didn’t.
- The higher degree models all fit and coefficients with high significance but constant and higher degree coefficients with very low significance.
These results are consistent with the LOOCV error results, which suggested a second degree model was best. If we decide which predictors to reject based on these hypothesis tests, we would end up with a model
which is the form of the true model