islr notes and exercises from An Introduction to Statistical Learning
3. Linear Regression
Exercise 10: Multiple regression of Sales on Price, Urban, and US features in Carseats dataset
- Preparing the dataset
- a. Fitting the model
- b. Interpreting the coefficients
- c. The formal model
- d. Significant Variables
- e. Fitting a second model with
price,USfeatures - f. Goodness-of-fit for the two models
- g. Confidence intervals for coefficients in the second model
- h. Outliers and high leverage points in the second model
- Footnotes
Preparing the dataset
{
The Carseats dataset is part of the ISLR R package. We’ll use rpy2 to import ISLR into python
import numpy as np
import pandas as pd
carseats = pd.read_csv('../../datasets/Carseats.csv')
We’ll check for null entries
carseats.isna().sum().sum()
0
a. Fitting the model
Both Urban and US are binary class variables, so we can represent both by indicator variables (see section 3.3.1.
To encode the qualitative variables Urban and US, we’ll use a LabelEncoder from sklearn
from sklearn.preprocessing import LabelEncoder
# instantiate and encode labels
urban_le, us_le = LabelEncoder(), LabelEncoder()
urban_le.fit(carseats.Urban.unique())
us_le.fit(carseats.US.unique())
LabelEncoder()
urban_le.classes_
array(['No', 'Yes'], dtype=object)
Now we’ll create a dataframe with the qualitative variables numerically encoded
# copy
carseats_enc = carseats.copy()
# transform columns
carseats_enc.loc[ :, 'Urban'] = urban_le.transform(carseats['Urban'])
carseats_enc.loc[ :, 'US'] = us_le.transform(carseats['US'])
carseats_enc[['Urban', 'US']].head()
| Urban | US | |
|---|---|---|
| 0 | 1 | 1 |
| 1 | 1 | 1 |
| 2 | 1 | 1 |
| 3 | 1 | 1 |
| 4 | 1 | 0 |
Now we can fit the model
import statsmodels.formula.api as smf
model = smf.ols('Sales ~ Urban + US + Price', data=carseats_enc).fit()
model.summary()
| Dep. Variable: | Sales | R-squared: | 0.239 |
|---|---|---|---|
| Model: | OLS | Adj. R-squared: | 0.234 |
| Method: | Least Squares | F-statistic: | 41.52 |
| Date: | Sun, 11 Nov 2018 | Prob (F-statistic): | 2.39e-23 |
| Time: | 15:41:28 | Log-Likelihood: | -927.66 |
| No. Observations: | 400 | AIC: | 1863. |
| Df Residuals: | 396 | BIC: | 1879. |
| Df Model: | 3 | ||
| Covariance Type: | nonrobust |
| coef | std err | t | P>|t| | [0.025 | 0.975] | |
|---|---|---|---|---|---|---|
| Intercept | 13.0435 | 0.651 | 20.036 | 0.000 | 11.764 | 14.323 |
| Urban | -0.0219 | 0.272 | -0.081 | 0.936 | -0.556 | 0.512 |
| US | 1.2006 | 0.259 | 4.635 | 0.000 | 0.691 | 1.710 |
| Price | -0.0545 | 0.005 | -10.389 | 0.000 | -0.065 | -0.044 |
| Omnibus: | 0.676 | Durbin-Watson: | 1.912 |
|---|---|---|---|
| Prob(Omnibus): | 0.713 | Jarque-Bera (JB): | 0.758 |
| Skew: | 0.093 | Prob(JB): | 0.684 |
| Kurtosis: | 2.897 | Cond. No. | 628. |
Warnings:
[1] Standard Errors assume that the covariance matrix of the errors is correctly specified.
b. Interpreting the coefficients
model.params
Intercept 13.043469
Urban -0.021916
US 1.200573
Price -0.054459
dtype: float64
Carseats has carseat sales data for 400 stores 1
can interpret these coefficients as follows
- The intercept is the (estimated/predicted) average sales for carseats sold for free (
Price=0) outside the US (US='No') and outside a city (Urban='No'). - is the estimated average sales for carseats sold in cities outside the US
- is the estimated average sales for carseats sold outside cities in the US
- is the estimated average sales for carseats sold in cities in the US
- so a $1 increase in price is estimated to decrease sales by
c. The formal model
Here we write the model in equation form. Let be Urban, be US, be Price. Then the model is
For the -th observation, we have
d. Significant Variables
is_stat_sig = model.pvalues < 0.05
model.pvalues[is_stat_sig]
Intercept 3.626602e-62
US 4.860245e-06
Price 1.609917e-22
dtype: float64
So we reject the null hypothesis for , that is for the variables US and price.
model.pvalues[~ is_stat_sig]
Urban 0.935739
dtype: float64
We fail to reject the null hypothesis for , that is for the variables Urban
e. Fitting a second model with price, US features
On the basis of d., we fit a new model with only the price and US variables
model_2 = smf.ols('Sales ~ US + Price', data=carseats_enc).fit()
model_2.summary()
| Dep. Variable: | Sales | R-squared: | 0.239 |
|---|---|---|---|
| Model: | OLS | Adj. R-squared: | 0.235 |
| Method: | Least Squares | F-statistic: | 62.43 |
| Date: | Sun, 11 Nov 2018 | Prob (F-statistic): | 2.66e-24 |
| Time: | 15:41:28 | Log-Likelihood: | -927.66 |
| No. Observations: | 400 | AIC: | 1861. |
| Df Residuals: | 397 | BIC: | 1873. |
| Df Model: | 2 | ||
| Covariance Type: | nonrobust |
| coef | std err | t | P>|t| | [0.025 | 0.975] | |
|---|---|---|---|---|---|---|
| Intercept | 13.0308 | 0.631 | 20.652 | 0.000 | 11.790 | 14.271 |
| US | 1.1996 | 0.258 | 4.641 | 0.000 | 0.692 | 1.708 |
| Price | -0.0545 | 0.005 | -10.416 | 0.000 | -0.065 | -0.044 |
| Omnibus: | 0.666 | Durbin-Watson: | 1.912 |
|---|---|---|---|
| Prob(Omnibus): | 0.717 | Jarque-Bera (JB): | 0.749 |
| Skew: | 0.092 | Prob(JB): | 0.688 |
| Kurtosis: | 2.895 | Cond. No. | 607. |
Warnings:
[1] Standard Errors assume that the covariance matrix of the errors is correctly specified.
f. Goodness-of-fit for the two models
One way of determining how well the models fit the data is the $R^2$ values
print("The first model has R^2 = {}".format(round(model.rsquared, 3)))
print("The second model has R^2 = {}".format(round(model_2.rsquared, 3)))
The first model has R^2 = 0.239
The second model has R^2 = 0.239
So as far as the value is concerned, the models are equivalent. To choose one over the other, we might look at prediction accuracy, comparing mean squared errors, for example
print("The first model has model mean squared error {}".format(round(model.mse_model, 3)))
print("The first model has residual mean squared error {}".format(round(model.mse_resid, 3)))
print("The first model has total mean squared error {}".format(round(model.mse_total, 3)))
The first model has model mean squared error 253.813
The first model has residual mean squared error 6.113
The first model has total mean squared error 7.976
print("The second model has model mean squared error {}".format(round(model_2.mse_model, 3)))
print("The second model has residual mean squared error {}".format(round(model_2.mse_resid, 3)))
print("The second model has total mean squared error {}".format(round(model_2.mse_total, 3)))
The second model has model mean squared error 380.7
The second model has residual mean squared error 6.098
The second model has total mean squared error 7.976
print("The second model's model mse is {}% of the first".format(
round(((model_2.mse_model - model.mse_model) * 100) / model.mse_model, 3)))
print("The second model's residual mse is {}% of the first".format(
round(((model_2.mse_resid - model.mse_resid) * 100) / model.mse_resid, 3)))
print("The second model's total mse is {}% of the first".format(
round(((model_2.mse_total - model.mse_total) * 100) / model.mse_total, 3)))
The second model's model mse is 49.992% of the first
The second model's residual mse is -0.25% of the first
The second model's total mse is 0.0% of the first
The statsmodel documentation has descriptions of these mean squared errors2.
- The model mean squared error is “the explained sum of squares divided by the model degrees of freedom”
- The residual mean squared error is “the sum of squared residuals divided by the residual degrees of freedom”
- The total mean squared error is “the uncentered total sum of squares divided by n the number of observations”
In OLS multiple regression with observations and predictors, the number of model degrees of freedom are , the number of residual degrees of freedom are , so we have
From this we can draw some conclusions
- It’s clear why for the two models in this problem
- The error is slightly smaller () for the second model, so it’s slightly at predicting the response.
- The error is much larger () for the second model, so for this model the response predictions have a much greater deviation from average
From this perspective, the first model seems preferable, but it’s difficult to say. The tiebreaker should come from their performance on test data.
g. Confidence intervals for coefficients in the second model
model_2.conf_int()
| 0 | 1 | |
|---|---|---|
| Intercept | 11.79032 | 14.271265 |
| US | 0.69152 | 1.707766 |
| Price | -0.06476 | -0.044195 |
h. Outliers and high leverage points in the second model
To check for outliers we look at a standardized residuals vs fitted values plot
import matplotlib.pyplot as plt
import seaborn as sns
%matplotlib inline
sns.regplot(model_2.get_prediction().summary_frame()['mean'], model_2.resid/model_2.resid.std(),
lowess=True,
line_kws={'color':'r', 'lw':1},
scatter_kws={'facecolors':'grey', 'edgecolors':'grey', 'alpha':0.4})
plt.xlabel('fitted values')
plt.ylabel('studentized resid')
Text(0,0.5,'studentized resid')
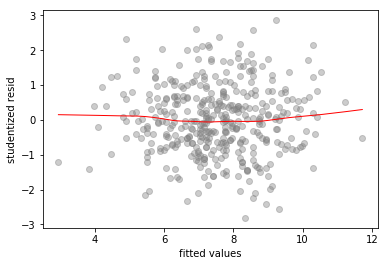
No outliers – all the studentized residual values are in the interval
((model_2.resid/model_2.resid.std()).min(), (model_2.resid/model_2.resid.std()).max())
(-2.812135116227541, 2.8627420897506766)
To check for high influence points we do an influence plot
# scatterplot of leverage vs studentized residuals
axes = sns.regplot(model_2.get_influence().hat_matrix_diag, model_2.resid/model_2.resid.std(),
lowess=True,
line_kws={'color':'r', 'lw':1},
scatter_kws={'facecolors':'grey', 'edgecolors':'grey', 'alpha':0.4})
plt.xlabel('leverage')
plt.ylabel('studentized resid')
# plot Cook's distance contours for D = 0.5, D = 1
x = np.linspace(axes.get_xlim()[0], axes.get_xlim()[1], 50)
plt.plot(x, np.sqrt(0.5*(1 - x)/x), color='red', linestyle='dashed')
plt.plot(x, np.sqrt((1 - x)/x), color='red', linestyle='dashed')
[<matplotlib.lines.Line2D at 0x1a181ff6a0>]
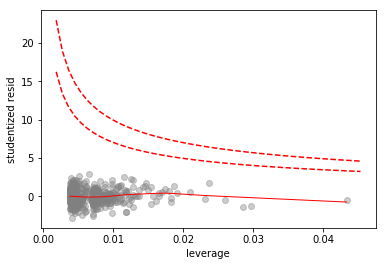
No high influence points. There is one rather high leverage point, but it has a low residual.